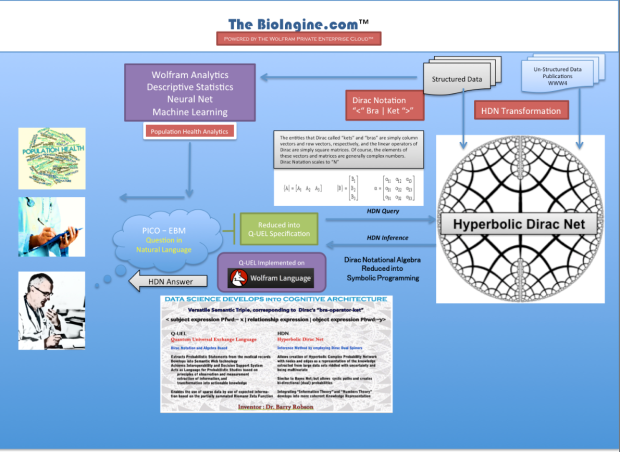
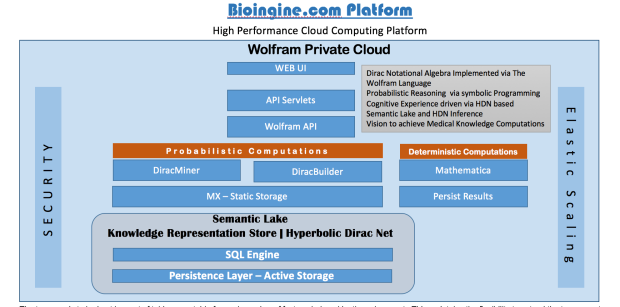
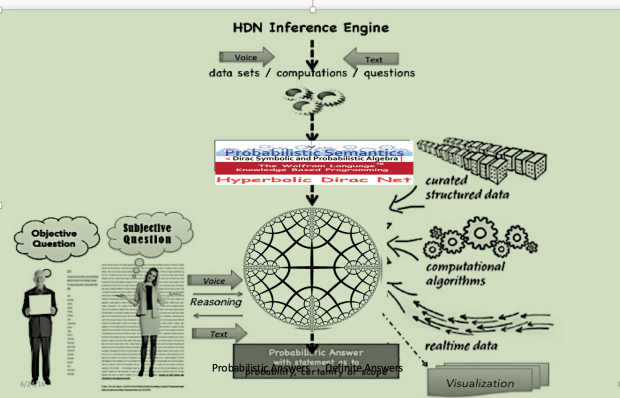
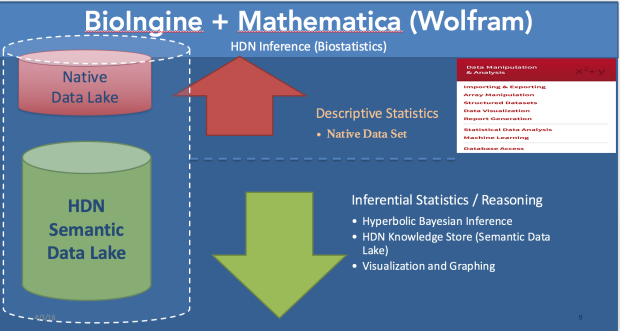
The BioIngine.com Platform Beta launch on the anvil with below discussed EBM examples for all to Explore !!!
The Bioingine.com Platform is built on Wolfram Enterprise Private Cloud
- using the technology from one of the leading science and tech companies
- using Wolfram Technology, the same technology that is at every Fortune 500 company
- using Wolfram Technology, the same technology that is at every major educational facility in the world
- leveraging the same technology as Wolfram|Alpha, the brains behind Apple’s Siri
Medical Automated Reasoning Programming Language environment [MARPLE]
References:- On PICO Gold Standard
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3140151/
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1069073/
Formulating a researchable question: A critical step for facilitating good clinical research
Sadaf Aslam and Patricia Emmanuel
Abstract:- Developing a researchable question is one of the challenging tasks a researcher encounters when initiating a project. Both, unanswered issues in current clinical practice or when experiences dictate alternative therapies may provoke an investigator to formulate a clinical research question. This article will assist researchers by providing step-by-step guidance on the formulation of a research question. This paper also describes PICO (population, intervention, control, and outcomes) criteria in framing a research question. Finally, we also assess the characteristics of a research question in the context of initiating a research project.
Keywords: Clinical research project, PICO format, research question
MARPLE – Question Format Medical Exam / PICO Setting
A good way to use Marple/HDNsudent is to set it up like an exam then the student answers. Marple then answers with its choices, i.e. candidate answers ranked by probability proposing its own choice of answer as the most probable and explaining why it did that (by the knowledge elements successfully used). This can then be compared with the intended answer of the examiner of which, of course Marple’s probability assessment of it can be seen.
It is already the case that MARPLE is used to test exam questions and it is scary that questions that have been issued by a Medical Licensing Board can turn out to be assigned an incorrect or unreachable answer by the examiner. The reason on inspection is that the question was ambiguous and potentially misleading, even though that may have not been obvious, or simply out of date – progress in science changed the answer and it shows up fast on some new web page (Translational Research for Medicine in action!). Often it is wrong or misleading because there turns out to be a very strong alternative answer.

Formulating the Questions in PICO Format
The modern approach to formulation is the recommendation for medical best practice known as PICO.
- P is the patient, population or problem (Primarily, what is the disease/diagnosis Dx?)
- I is intervention or something happening that intervenes (What is the proposed therapy Rx (drug, surgery, or life style recommendation)
- C is some alternative to that intervention or something happening that can be compared (with what options (including no treatment)? May also include this in the context of different compared types of patient female, diabetic, elderly, or Hispanic etc.
- O is the outcome, i.e. a disease state or set of such that occurs, or fails to occur, or is ideally terminated by the intervention such that health is restored. (Possibly that often means the prognosis, but often prognosis implies a more complex scenario on a longer timescale further in the future).
Put briefly “For P does I as opposed to C have outcome O” is the PICO form.
The above kinds of probabilities are not necessarily the same as an essentially statistical analysis by structured data mining would deliver. All of these except C relate to associations, symptoms, Dx, Rx, outcome. It is C that is difficult. Probably the best interpretation is replacing Rx in associations with no Rx and then various other Rx. If C means say in other kinds of patients, then it is a matter of associations including those.
A second step of quantification is usually required in which probabilities are obtained as measures of scope based on counting. Of particular interest here is the odds ratio
Two Primary Methods of Asking a Question in The BioIngine
1. Primarily Symbolic and Qualitative. (more unstructured data dependent) [Release 1]
HDN is behind the scenes but focuses mainly on contextual probabilities between statements. HDNstudent is used to address the issue as a multiple choice exam with indefinitely large numbers of candidate answers, in which the expert end-user can formulate PICO questions and candidate answers, or all these can be derived automatically or semi-automatically. Each initial question can be split into a P, I, C, and O question.
2. Primarily Calculative and Quantitative. (more structured – EHR data dependent) [Release 2]
Focus on intrinsic probabilities, the degree of truth associated with each statement by itself. DiracBuilder used after DiracMiner addresses EBM decision measures as special cases of HDN inference. Of particular interest is an entry
<O | P, I > / <O | P, C>
which is the HDN likelihood or HDN relative risk of the outcome O given patient/population/problem P given I as opposed to C, usually seen as a “NOT I”, and
<NOT O | P, I> / <NOT O | P, C>
which is the HDN likelihood or HDN relative risk of NOT getting the outcome O given patient/population/problem P given I as opposed to C usually seen as a “NOT I”. Note though that you get a two for one, because we also have <P, I | O>, the adjoint form, at the same time, because on the complex conjugate of the other. Note that the ODDS RATIO is the former likelihood ratio over the latter, and hence the HDN odds ratio as it would normally be entered in DiracBuilder is as follows:-
<O | P, I>
/<NOT O | P, C>
<NOT O | P, C>
/<NOT O | P, I>
- QUALITATIVE / SYMBOLIC:
“An 84-year-old man in a nursing home has increasing poorly localized lower abdominal pain recurring every 3-4 hours over the past 3 days. He has no nausea or vomiting; the last bowel movement was not recorded. Examination shows a soft abdomen with a palpable, slightly tender, lower left abdominal mass. Hematocrit is 28%. Leukocyte count is 10,000/mm3. Serum amylase activity is within normal limits. Test of the stool for occult blood is positive. What is the diagnosis?”
•This is usually addressed by a declared list of multiple choice candidate answers, though the list can be indefinitely large. 30 is not unusual.
•The answers are all assigned probabilities, and the most probable is considered the answer, at least for testing purposes in a medical licensing exam context. These probabilities can make use of probabilities, but predominantly they are contextual probabilities, depending in the relationships between chains and networks of knowledge elements that link the question to each answer.
- QUANTITATIVE / CALCULATIVE:
“Will my female patient age 50-59 taking diabetes medication and having a body mass index of 30-39 have very high cholesterol if the systolic BP is 130-139 mmHg and HDL is 50-59 mg/dL and non-HDL is 120-129 mg/dL?”.
•This forms a preliminary Hyperbolic Dirac Net (inference net) from the query, which may be refined and to each statement intrinsic probabilities are assigned, e.g. automatically by data mining.
•This question could properly start “What is the probability that…” . The real answers of interest here are not qualitative statements, but the final probabilities.
•Note the “IF”. But POPPER extends this to relationships beyond IF associative or conditional ones, e.g. verbs of action.
Quantitative Computations :- Odds Ratio and Risk Computations
- Medical Necessity
- Laboratory Testing Principles
- Quality of Diagnosis
- Diagnosis Test Accuracy
- Diagnosis Test
- Sensitivity
- Specificity
- Predictive Values – Employing Bayes Theorem (Positive and Negative Value)
- Coefficient of Variations
- Resolving Power
- Prevalence and Incidence
- Prevalence and Rate
- Relative Risk and Cohort Studies
- Predictive Odds
- Attributable Risk
- Odds Ratio
Examples Quantitative / Calculative HDN Queries
In The Bioingine.com Release 1 – we are only dealing with Quantitative / Calculative type questions
Examples discussed in section A below are simple to play with to appreciate the HDN power for conducting inference. However, Problems B2 onwards requires some deeper understanding of the Bayesian and HDN analysis.
<‘Taking BP medication’:=’1’ | ‘Taking diabetes medication’:= ‘1’>
/<‘Taking BP medication’:=’1’ | ‘Taking diabetes medication’:= ‘0’>
A. Against Data Set 1.csv (2114 records with 33 variables created for Cardiovascular Risk Studies (Framingham Risk Factor)
B. Against Data Set2.csv (nearing 700,000 records with 196 variables. Truly a large data set with high dimensionality (many columns of clinical and demographic factors), leading to a combinatorial explosion.
Note: in the examples below, you are forming questions or HDN queries such as
“For African Caribbean patients 50-59 years old with a BMI of 50-59 what is the Relative Risk of needing to be on BP medication if there is a family history as opposed to no family history?
IMPORTANT: THE TWO-FOR-ONE EFFECT OF THE DUAL. Calculations report a dual value for any probabilistic value implied for the expression ented. In some cases you may be only interest in the first number in the dual, but the second number is always meaningful and frequently very useful. Notably, we say Relative Risk by itself for brevity, but in fact this is only the first number in the dual that is reported. In general, the form
<’A’:=’1’|’B’:=’1’>
/<’A’:=’1’|’B’:=’0’>
yields the following dual probabilistic value…
(P(’A’:=’1’|’B’:=’1’)/ P(’A’:=’1’|’B’:=’0’), ( P(’B’:=’1’|’A’:=’1’)/ P(’B’:=0’|’B’:=’1’),
where the first ratio is relative risk RR(P(’A’:=’1’|’B’:=’1’) and the second ratio is predictive odds RR(P(’A’:=’1’|’B’:=’1’).
a. This inquiry seeking the risk of BP requires being translated into Q-UEL specification as shown below. [All the below Q-UEL queries in red can be copied and entered in the HDN query to get the HDN inference for the pertinent Data Sets.]
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1 ‘ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and BMI:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’0’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
b. The Q-UEL specified query enables Notational Algebra to work while making inference from the giant semantic lake or the knowledge repository store (KRS).
c. Recall, KRS is the representation of the universe as a Hyperbolic Dirac Net. This was created by transformation process of the uploaded data set to activate the automated statistical studies.
d. The query works against the KRS and extracts the inference in HDN format displaying an inverse Bayesian Result; which calculates both classical and zeta probabilities :- Pfwd, Pzfwd & Pbwd, Pzbwd
A1. Relative Risk – High BP Case
Example: – Study of BP = blood pressure (high) in the population data set considered.
This case is very similar, because high BP and diabetes are each comorbidities with high BMI and hence to some extent with each other. Consequently we just substitute diabetes by BP throughout.
Note: for the values enter discreet or continuous
(0) We can in fact test the strength of the above with the following RR, which in effect reads as “What is the relative risk of needing to take BP medication if you are diabetic as opposed to not diabetic?
<‘Taking BP medication’:=’1’ | ‘Taking diabetes medication’:= ‘1’>
/<‘Taking BP medication’:=’1’ | ‘Taking diabetes medication’:= ‘0’>
The following predictive odds PO make sense and are useful here:-
<‘Taking BP medication’:=’1’ | ‘BMI’:= ’50-59’ >
/<‘Taking BP medication’:=’0’ | ‘BMI’:= ’50-59’ >
and (separately entered)
<‘Taking diabetes medication’:=’1’ | ‘BMI’:= ’50-59’ >
/<‘Taking diabetes medication’:=’0’ | ‘BMI’:= ’50-59’ >
And the odds ratio OR would be a good measure here (as it works in both directions). Note Pfwd = Pbw theoretically for an odds ratio.
<‘Taking BP medication’:=’1’ | ‘Taking diabetes medication’:= ‘1’>
<‘Taking BP medication’:=’0’ | ‘Taking diabetes medication’:= ‘0’>
/<‘Taking BP medication’:=’1’ | ‘Taking diabetes medication’:= ‘0’>
/<‘Taking BP medication’:=’0’ | ‘Taking diabetes medication’:= ‘1’>
(1) For African Caribbean patients 50-59 years old with a BMI of 50-59 what is the Relative Risk of needing to be on BP medication if there is a family history as opposed to no family history?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1‘ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and BMI:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’0’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
(2) For African Caribbean patients 50-59 years old with a family history of BP what is the Relative Risk of needing to be on BP medication if there is a BMI of 50-59 as opposed to a reasonable BMI of ’20-29’?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’20-29’ >
(3) For African Caribbean patients with a family history of BP, what is the Relative Risk of needing to be on BP medication if there is an age of 50-59 rather than 40-49?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’40-49’ and ‘BMI’:= ’50-59’>
(4) For African Caribbean patients with a family history of BP, what is the Relative Risk of needing to be on BP medication if there is an age of 50-59 rather than 40-49?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59and ‘BMI’:= ’40-49’>
(5) For African Caribbean patients with a family history of BP, what is the Relative Risk of needing to be on BP medication if there is an age of 50-59 rather than 40-49?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1‘ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59and ‘BMI’:= ’40-49’>
(6) For African Caribbean patients with a family history of BP, what is the Relative Risk of needing to be on BP medication if there is an age of 50-59 rather than 30-39?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’30-39and ‘BMI’:= ’40-49’>
(7) For African Caribbean patients with a family history of BP, what is the Relative Risk of needing to be on BP medication if there is an age of 50-59 rather than 20-29?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’20-29 and ‘BMI’:= ’40-49’>
(8) For patients with a family history of BP age 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on BP medication if they are African Caribbean rather than Caucasian?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘Caucasian’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
(9) For patients with a family history of BP age 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on BP medication if they are African Caribbean rather than Asian?
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’’1 and ‘Ethnicity’:=‘Asian’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
(10) For patients with a family history of BP age 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on BP medication if they are African Caribbean rather than Hispanic
< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking BP medication’:=’1’ | ‘Family history of BP’:=’1’ and ‘Ethnicity’:=‘Hispanic’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
A2. Relative Risk – Diabetes Case
Against Data Set1.csv
Type 2 diabetes is implied here.
(11) For African Caribbean patients 50-59 years old with a BMI of 50-59 what is the Relative Risk of needing to be on diabetes medication if there is a family history as opposed to no family history?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and BMI:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’0’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
(12) For African Caribbean patients 50-59 years old with a family history of diabetes what is the Relative Risk of needing to be on diabetes medication if there is a BMI of 50-59 as opposed to a reasonable BMI of ’20-29’?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’20-29’ >
(13) For African Caribbean patients with a family history of diabetes, what is the Relative Risk of needing to be on diabetes medication if there is an age of 50-59 rather than 40-49?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’40-49’ and ‘BMI’:= ’50-59’>
(14) For African Caribbean patients with a family history of diabetes, what is the Relative Risk of needing to be on diabetes medication if there is an age of 50-59 rather than 40-49?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59and ‘BMI’:= ’40-49’>
(15) For African Caribbean patients with a family history of diabetes, what is the Relative Risk of needing to be on diabetes medication if there is an age of 50-59 rather than 40-49?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59and ‘BMI’:= ’40-49’>
(16) For African Caribbean patients with a family history of diabetes, what is the Relative Risk of needing to be on diabetes medication if there is an age of 50-59 rather than 30-39?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’30-39and ‘BMI’:= ’40-49’>
(17) For African Caribbean patients with a family history of diabetes, what is the Relative Risk of needing to be on diabetes medication if there is an age of 50-59 rather than 20-29?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’20-29and ‘BMI’:= ’40-49’>
A3. Relative Risk – Cholesterol Case
Against Data Set1.csv
(18) For African Caribbean patients 50-59 years old with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if there is a family history as opposed to no family history?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and BMI:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
(19) For African Caribbean patients 50-59 years old with a fat% of 40-49, with a family history of cholesterol, what is the Relative Risk of needing to be on cholesterol medication if there is a BMI of 50-59 as opposed to a reasonable BMI of ’20-29’?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’20-29’ >
(20) For African Caribbean patients with a family history of cholesterol, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if there is an age of 50-59 rather than 40-49?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’40-49’ and ‘BMI’:= ’50-59’>
(21) For African Caribbean patients with a family history of cholesterol, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if there is an age of 50-59 rather than 40-49?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59and ‘BMI’:= ’40-49’>
(22) For African Caribbean patients with a family history of cholesterol, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if there is an age of 50-59 rather than 40-49?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59and ‘BMI’:= ’40-49’>
(23) For African Caribbean patients with a family history of cholesterol , with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if there is an age of 50-59 rather than 30-39?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’30-39and ‘BMI’:= ’40-49’>
(24) For African Caribbean patients with a family history of cholesterol, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if there is an age of 50-59 rather than 20-29?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’20-29and ‘BMI’:= ’40-49’>
(25) For patients with a family history of cholesterol age 50-59 and BMI of 50-59, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if they are African Caribbean rather than Caucasian?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=1‘’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘Caucasian’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
(26) For patients with a family history of cholesterol age 50-59 and BMI of 50-59, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if they are African Caribbean rather than Asian?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘Asian’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
(27) For patients with a family history of cholesterol age 50-59 and BMI of 50-59, with a fat% of 40-49, what is the Relative Risk of needing to be on cholesterol medication if they are African Caribbean rather than Hispanic
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:=‘Hispanic’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
(28) For ‘African Caribbean’ patients with a family history of cholesterol age 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on cholesterol medication if they have fat% 40-49 rather than 30-39?
< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:= ‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking cholesterol medication’:=‘1’ | ‘Fat(%)’:=‘40-49’ and ‘Ethnicity’:= ‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59>
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘Caucasian’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’>
(29) For patients with a family history of diabetes age 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on diabetes medication if they are African Caribbean rather than Asian?
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘Asian’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’>
(30) For patients with a family history of diabetes age 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on diabetes medication if they are African Caribbean rather than Hispanic
< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘African Caribbean’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-59’ >
/< ‘Taking diabetes medication’:=’1’ | ‘Family history of diabetes’:=’1’ and ‘Ethnicity’:=‘Hispanic’ and ‘age(years):=’50-59’ and ‘BMI’:= ’50-5’9>
(31) For patients with a family history of diabetesage 50-59 and BMI of 50-59, what is the Relative Risk of needing to be on diabetes medication if they are African Caribbean rather than Caucasian?





























You must be logged in to post a comment.